标签:ali sig head ble name 细胞 bsp require data
# devtools::install_github("satijalab/seurat-data")
library(SeuratData)
# AvailableData()
# InstallData("pbmc3k.SeuratData")
data(pbmc3k)
exp <- pbmc3k@assays$RNA@data
dim(exp)
# exp[1:5,1:5]
table(is.na(pbmc3k$seurat_annotations))
table(pbmc3k$seurat_annotations)
library(Seurat)
pbmc3k@active.ident <- pbmc3k$seurat_annotations
table(pbmc3k@active.ident)
deg <- FindMarkers(pbmc3k, ident.1 = "Naive CD4 T", ident.2 = "B")
# head(deg)
dim(deg)
# if (!requireNamespace("BiocManager", quietly = TRUE))
# install.packages("BiocManager")
# BiocManager::install("GSEABase")
library(GSEABase)
library(ggplot2)
library(clusterProfiler)
library(org.Hs.eg.db)
# API 1
geneList <- deg$avg_logFC
names(geneList) <- toupper(rownames(deg))
geneList <- sort(geneList, decreasing = T)
head(geneList)
# API 2
# gmtfile <- "../EllyLab//human/singleCell/MsigDB/msigdb.v7.4.symbols.gmt"
gmtfile <- "../EllyLab//human/singleCell/MsigDB/c5.go.bp.v7.4.symbols.gmt"
geneset <- read.gmt(gmtfile)
length(unique(geneset$ont))
egmt <- GSEA(geneList, TERM2GENE = geneset, minGSSize = 1, pvalueCutoff = 0.99, verbose = F)
# head(egmt)
gsea.out.df <- egmt@result
# gsea.out.df
# head(gsea.out.df$ID)
library(enrichplot)
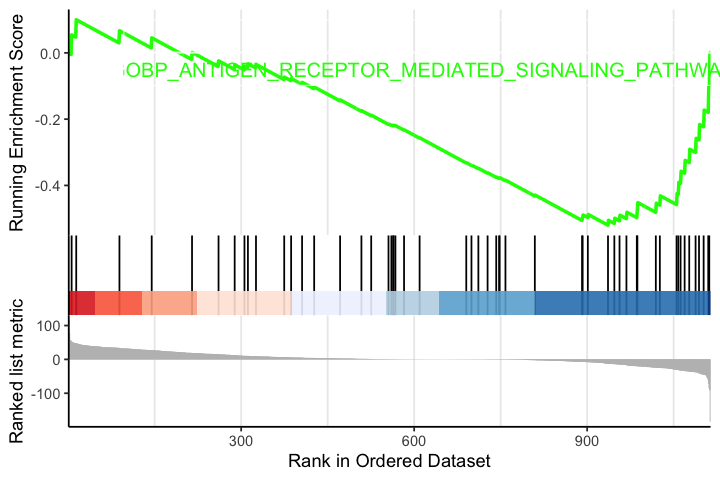
options(repr.plot.width=6, repr.plot.height=4) gseaplot2(egmt, geneSetID = "GOBP_ANTIGEN_RECEPTOR_MEDIATED_SIGNALING_PATHWAY", pvalue_table = T)
标签:ali sig head ble name 细胞 bsp require data
原文地址:https://www.cnblogs.com/leezx/p/14754812.html